Abstract
[Background]
CD34-positive monocytes (CD34+mono) have recently been identified following mobilization by granulocyte-colony stimulating factor (G-CSF), and have been suggested to have a potential to modulate immune functions in animal models. However, the biological feature of CD34+mono in humans still remains unclear. Thus, we explored the difference between CD34+mono, CD34+cells, and monocytes through the analyses of gene expression profiles (GEP).
[Methods]
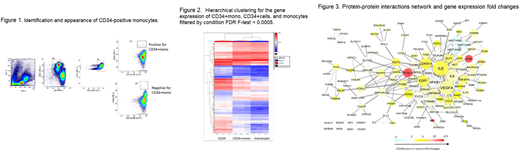
CD34+mono (Lin-CD34+CD33+CD14+CD11b+, Figure1), CD34+cells (Lin-CD34+CD33-CD14-CD11b- ), and monocytes (Lin-CD34-CD33+CD14+CD11b+) were directly sorted into tubes from cryopreserved grafts from three healthy donors. After the extraction of total RNA, microarray analyses were performed with GeneChip™ 3' IVT Pico Kit and analyzed according to the algorithm of Transcriptome Analysis Console (Thermo Fisher Scientific). Condition false discovery rate (FDR) F-test was used for filtering genes, and the threshold was <0.0005 for hierarchical clustering without considering fold changes. In addition, enrichment analyses were performed for gene ontology (GO) and pathways by the Kyoto Encyclopedia of Genes and Genomes (KEGG) through Database for Annotation, Visualization and Integrated Discovery (DAVID). The differentially expressed genes with <0.05 of F-test values and 2 or more fold changes in CD34+mono compared with CD34+cells and monocytes were selected for the enrichment analyses. Thereafter, the protein-protein interaction (PPI) network in CD34+mono was constructed through Search Tool for the Retrieval of Interacting Genes/Proteins (STRING), which is a biological database. This study was approved by the institutional review board of Jichi Medical University and all subjects gave their written informed consent for the cryopreservation and analysis of the blood samples in accordance with the Helsinki Declaration.
[Results]
CD34+mono resembled con-mono in the appearance, although it might look slightly immature. In fact, the number of differential expressed genes was smaller in the pair of CD34+mono vs. monocytes (n=605) compared with the other pair (n=2136), suggesting that CD34+mono might stand nearby monocytes. However, a principle component analysis of GEP demonstrated that CD34+mono would be categorized into an independent cell type entity. In addition, the hierarchical clustering heat map suggested that CD34+mono would have the interim gene expressions between CD34+ cells and monocytes (Figure 2).
GO analysis was also performed using the 203 genes which were differentially expressed in CD34+mono compared with CD34+cells and monocytes. The differentially expressed genes seemed to be significantly involved in the "immune system process" of biological process, followed by "locomotion", "metabolic process" and "response to stimulus", because pro-inflammatory genes in CD34+mono like IL6, CCL3, IL8, VEGFA, and IL1A were increased.
In addition, PPI analyses indicated that 128 of the 203 differentially expressed genes in CD34+mono had close connections with each other. Especially, IL6, VEGFA, IL8, NFkB1, EGR1, CDKN1A (p21), and CYCS could be considered as a hub gene in CD34+mono. Of them, IL6, IL8, and VEGFA are considered to be pro-inflammatory cytokines. NFkB1, and EGR1 are transcriptional factors, and CDKN1A, and CYCS are considered to regulate cell cycle or apoptosis. Furthermore, simultaneously focusing on the difference in the fold changes of gene expressions between CD34+mono vs. monocytes, CD83 (a membrane protein and immunoglobulin superfamily that regulates antigen presentation) and FOSL1 (a kind of regulators of cell proliferation, differentiation, and transformation) were specifically increased in CD34+mono (Figure3).
The increased expression of CD83 strongly suggests antigen presenting cells like mature dendritic cells. CD34+mono might be one of progenitors of dendritic cells, or might have a potential of antigen presentation itself.
[Conclusion]
In summary, the differentially expressed genes in CD34+mono would be mapped in the immune process, especially against infectious pathogens. In addition, the increased CD83 suggested that CD34+mono might play a role of antigen presentation in the immune response. Further investigations would be necessary to confirm the clinical roles of CD34+mono after transplantation.
Nakasone:Phizer: Honoraria; Novartis: Honoraria; Kyowa Hakko Kirin: Honoraria; Celgene: Honoraria; Bristol-Myers Squibb: Honoraria; Janssen: Honoraria; Takeda: Honoraria. Kimura:Astellas: Honoraria; Pfizer: Honoraria; Sumitomo Dainippon Pharma: Honoraria; MSD: Other: Investigator in the institute; Nippon Kayaku: Honoraria; Celgene: Honoraria; Kyowa Hakko Kirin: Honoraria; Takeda: Honoraria. Kako:Takeda Pharmaceutical Company Limited.: Honoraria; Takeda Pharmaceutical Company Limited.: Honoraria; Celgene K.K.: Honoraria; Bristol-Myers Squibb: Honoraria; Sumitomo Dainippon Pharma Co., Ltd.: Honoraria; Chugai Pharmaceutical Co., Ltd.: Honoraria; Otsuka Pharmaceutical Co., Ltd.: Honoraria; Ono Pharmaceutical Co., Ltd.: Honoraria; Janssen Pharmaceutical K.K.: Honoraria. Kanda:Eisai: Consultancy, Honoraria, Research Funding; Dainippon-Sumitomo: Consultancy, Honoraria, Research Funding; MSD: Research Funding; Nippon-Shinyaku: Research Funding; Pfizer: Research Funding; Kyowa-Hakko Kirin: Consultancy, Honoraria, Research Funding; Chugai: Consultancy, Honoraria, Research Funding; Otsuka: Research Funding; Taisho-Toyama: Research Funding; CSL Behring: Research Funding; Tanabe-Mitsubishi: Research Funding; Astellas: Consultancy, Honoraria, Research Funding; Bristol-Myers Squibb: Consultancy, Honoraria; Ono: Consultancy, Honoraria, Research Funding; Taiho: Research Funding; Shionogi: Consultancy, Honoraria, Research Funding; Sanofi: Research Funding; Takeda: Consultancy, Honoraria, Research Funding; Asahi-Kasei: Research Funding; Novartis: Research Funding; Celgene: Consultancy, Honoraria; Mochida: Consultancy, Honoraria; Alexion: Consultancy, Honoraria; Takara-bio: Consultancy, Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal